Applications
The LassoProt server/database can be used in various ways. First of all it can be used as a source of information about the topology and topological constraints in proteins that possess pierced closed loops [1-2]. Those constrains can have an impact on protein folding, unfolding, stability and function. Topological constrains can be further used to understand how proteins sustain harsh thermal and chemical conditions [3-4]. Moreover, a lot of information can be deduced from the results of trajectory analysis. Detected lassos have broad application as a reaction coordinate to follow folding and unfolding pathways not only of lasso proteins [3-4], but also of knotted proteins [5]. Furthermore, it can be applied to identify a mechanical clamp or disentangled topological glue during protein stretching [6] or to understand biological function of lasso when protein is exposed to different environments, e.g. oxidative stress. Moreover, topological fingerprint can be used as a template to identify proteins with desired properties, to identify “an unwanted” topology via e.g. engineering disulphides bonds, or to augment known structures. Finally, currently known lasso motifs can be used as a template to double check predicted protein structures in the CASP competition or to help to assign proper arrangement to the protein backbone in the ECM method or just in the X-ray.
Contents
- Analyzing trajectories, protein folding/unfolding, new reaction coordinate
- Stretching proteins, identification of mechanical clamps, untying complex topology
- Biological function
- Protein structure prediction
- Intelligent design of engineered disulphide bridges in proteins and other biopolymers, medicine
- Mathematics, detection of topology
[1] Niemyska W, Dabrowski-Tumanski P, Kadlof M, Haglund E, Sułkowski P, Sulkowska JI (2016) Complex lasso: new entangled motifs in proteins.
[2] Dabrowski-Tumanski P, Sulkowska JI (2016) Unique properties of lasso proteins. – under review
[3] Haglund E, Sulkowska JI, He Z, Feng GS, Jennings PA, Onuchic JN (2012) The unique cysteine knot regulates the pleotropic hormone leptin. PLoS One.7(9):e45654
[4] Haglund E, Sulkowska JI, Noel JK, Lammert H, Onuchic JN, Jennings PA (2014) Pierced Lasso Bundles are a new class of knot-like motifs. PLoS Comput Biol. Jun 19;10(6):e1003613
[5] Noel JK, Onuchic JN, Sulkowska JI (2013) Knotting a Protein in Explicit Solvent. J. Phys. Chem. Lett. 4 (21), pp 3570-3573
[6] Sikora M, Sulkowska JI, Cieplak M (2009) Mechanical strength of 17 134 proteins and cysteine slipknots. PLoS Comput Biol. 5(10):e1000547
[7] Haglund E (2015) Engineering covalent loops in proteins can serve as an on/off switch to regulate threaded topologies. J. Phys. Cond. Matter 27, 35
[8] 'The Knottin Database'
[9] Clark DP, Pazdernik NJ (2012) Biotechnology.
[10] Dombkowski AA (2003) Disulfide by design: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics 19 1852–3
[11] Silverman A P, Levin A M, Lahti J L and Cochran J R (2009) Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J. Mol. Biol. 385 1064–75
[12] Craik DJ, Simonsen S, Daly NL (2002) The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discovery Dev. 5 251–60
[13] Uehara E, Tanaka R, Inoue M, Hirose F, Deguchi T (2014) Mean-square radius of gyration and hydrodynamic radius for topological polymers evaluated through the quaternionic algorithm. Reactive and Functional Polymers, 80, 48–56
[14] Takuya Suzuki, Takuya Yamamoto, Yasuyuki Tezuka (2014) Constructing a Macromolecular K3,3 Graph through Electrostatic Self-Assembly and Covalent Fixation with a Dendritic Polymer Precursor. J. Am. Chem. Soc., 2014, 136 (28), pp 10148–10155
Analyzing trajectories, protein folding/unfolding, new reaction coordinate
Our server provides new tools for analyzing and characterizing trajectories. These tools can be used to better understand the free energy landscape of lasso proteins. For example, the results of the “fast analysis” (existence of a lasso at each time step) can be used to characterize the probability of lasso occurrence to support other reaction coordinates, such as Q (the fraction of native contacts detected in a protein at a given time step) or RMSD. As an example, Fig. 1 below shows folding pathways of the lasso protein under reduced/oxidized condition, described by Q (figure from the paper [3]). Although at the same value of Q proteins can show several similar configurations (the same protein in reduced and oxidized state), they possess different topology. Detected topological barrier explains the difference in the folding time under reducing/oxidizing conditions.
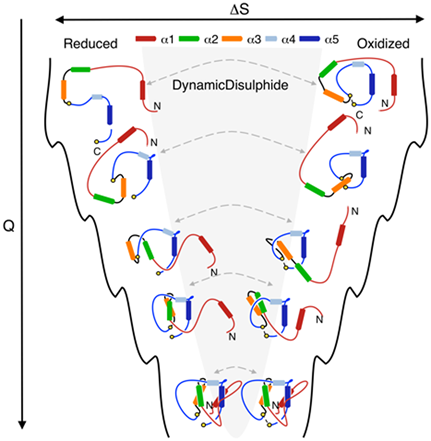
Fig. 1 Cartoon representation of the folding landscape of leptin, protein with L1 topology under reducing (left) and oxidizing (right) conditions, obtained in molecular dynamics simulation. Analysis of lasso topology is used here to explain the difference in folding time under reducing/oxidizing condition when proteins possess similar number of native contacts. Figure from the paper [3].
Yet another application amounts to describing knotting mechanism of entangled proteins or any other mechanism, which requires threading of one chain through some loop. The server allows the user to establish a closed loop via any type of interaction (e.g. a hydrogen bond). This option can be used to close a twisted loop of a knotted protein and establish a new reaction coordinate, “the loop crossing coordinate”, as discussed in [5]. The loop crossing coordinate can be used to analyze e.g. knotting via a slipknot configuration from the viewpoint of thermodynamics process and estimate effective potential, PMF. Based on such analysis, the figure like this below Fig. 2 can be plotted [5].
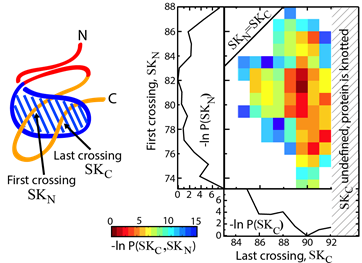
Fig. 2 Estimated effective potential (PMF) along the loop crossing coordinate for the smallest knotted protein (pdb code 2EFV). The left panel shows a schematic representation of a surface spanned on a twisted loop of the knotted protein, which is pierced via the C terminus twice (double lasso - L2). The right panel shows tracked behavior of residues during crossing the plane of the loop based on the explicit-solvent simulation [5]. The crossing data are used to construct an effective potential to describe knotting mechanism.
Another application arises from the analysis of the minimal surface: its dynamics and area fluctuations can serve as another, new reaction coordinate in complex protein folding. We would like to stress, that due to the possibility of analyzing a minimal surface spanned on an arbitrary loop, this reaction coordinate can be applied to any imaginary loop. Moreover, such reaction coordinate can be combined with others well known reaction coordinates, such as Q or RMSD, as in the Fig. 2. The information about the minimal surface and in particular about its area is available also for each protein deposited in LassoProt (see Fig. 3).
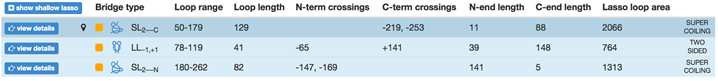
Fig. 3 Example of the protein with the complex lasso and assigned the “lasso loop area”.
Moreover, LassoProt server can be used to disentangle complex structure of the protein, obtained e.g. during simulations. The protein with any type of closed loop, or an ensemble of proteins with any type of closed loops, can form a complex structure of “topological glue”, which cannot be easily disentangled, although the loops are not joined (i.e. do not form a link) (see Fig. 4). However, it can be hard to distinguish “by naked eye” if such a situation occurred. Nonetheless, it can be easily identified with the topological information provided by our server.

Fig. 4 Two loops topologically glued together.
Stretching proteins, identification of mechanical clamps, untying complex topology
The information about the existence of topological glue or other complex structures is also important during analysis of protein stretching e.g. by AFM, optical tweezers, or by computer techniques. Firstly, observe that the loops in Fig. 4 can be easily separated if only they are pulled in one specific direction. If one pulls from the other side the loops get tied. Secondly, the server can be used to detect mechanical clamps or new topological motifs in proteins, which appear under mechanical tension. The Fig. 5 (from [6]) shows unraveling of the protein and identification of cystein slipknot motif during protein stretching with constant speed. Appearance of this motif corresponds to a mechanical clamp, which explains surprisingly high mechanical resistance, as discussed in [6].
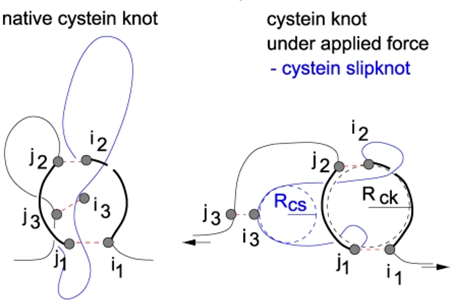
Fig. 5 Geometry of a cystein slipknot. The left figure shows a schematic representation of the native geometry of part of proteins that yields the cystein slipknot on stretching. The resulting cystein slipknot motif is shown in the right figure. The figure from [6].
Biological function
As yet another application, theoretical analysis of a lasso protein in different environments can lead to understanding of biological activity of this protein. E.g. proteins can be exposed to the oxidative stress [8], which leads to formation or breaking of cysteine bonds. Analysis of molecular simulations from the viewpoint of lasso topology (as shown in the Fig. 6) led to understanding experimental data about leptin, showing that lasso not only stabilizes a protein, but also changes the malleability of the native state, and thus leads to a change in biological activity.
We stress, that the complex lasso proteins are more general structures than cysteine knots, as cysteine knot requires the existence of (at least) three covalent loops. Therefore the analysis provided by us can be used to further characterize, or easily distinguish proteins with cysteine knots, as these should have a specified topology. Moreover, the crossing residue analysis can lead to explanation of stability properties of some proteins. E.g. miniproteins, being therapeutically relevant, possess lasso topology, which is stabilized by the existence of bulky residue in a vicinity of the crossing. The server predicts accurately the blocking residue, as shown in the Fig. 7. Modification of this residue can change properties of the protein. Detected blocking residues for 13 mini-proteins are available in [1].
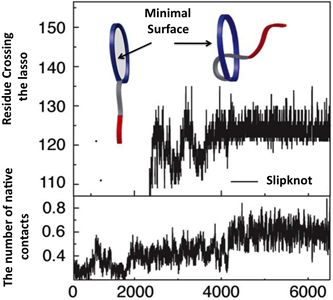
Fig. 6 The threading mechanism for the protein with the lasso motif L1. The upper panel shows the schematic representation of the protein without lasso L0 and with lasso motif L1 (right) based on a detected number of residue, which pierces through the N-terminal loop. This mechanism corresponds to the slipknoting via the C-terminal, the figure from [4].
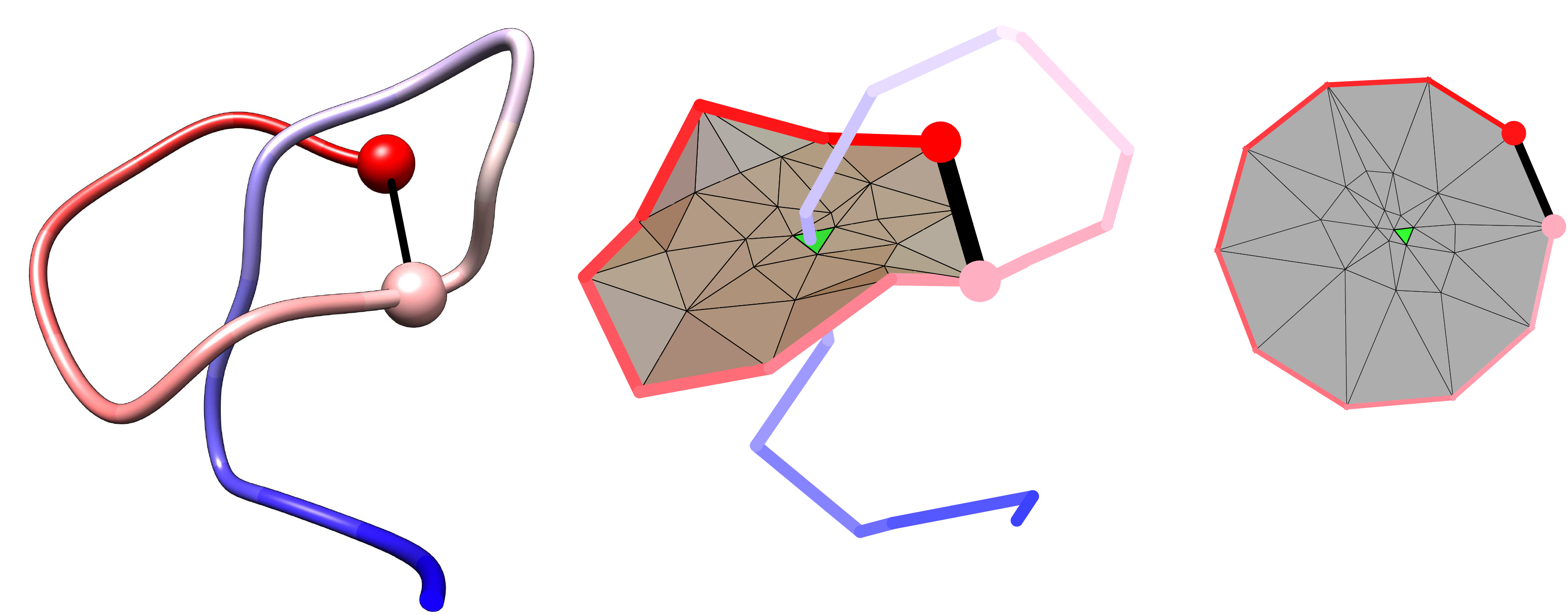
Fig. 7 Example structure of the miniprotein (pdb code 1RPB) with accurately detected blocking residue, determined by our server.
Protein structure prediction
Yet another application of the server is as a method to validate the structure of predicted proteins e.g. in the CASP competition or to double check the protein structure determined in ECM method experimentally. Currently all known lasso motifs (based on all proteins deposited in pdb) are listed in the lasso fingerprint table. Thus all new topological motifs predicted in the CASP competition should be evaluated carefully. Fig. 8 and 9 show respectively the proteins with correctly predicted lasso motif and proteins, which have high GDT score, but have a wrong lasso topology (even the topology that has not been identified in natural proteins yet, such as L3,1 and LS4).
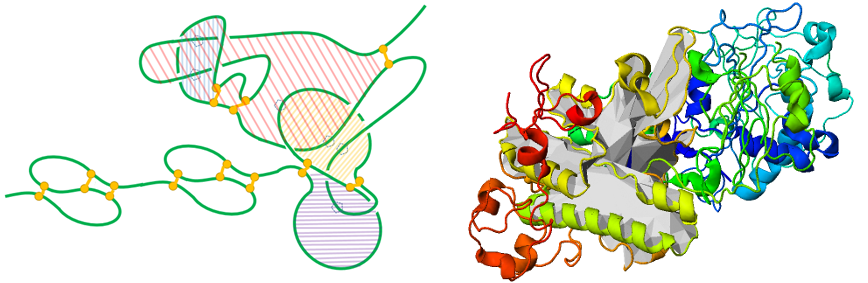
Fig. 8 Left panel - schematic representation of the complex lasso correctly predicted in the protein (CASP9) and carton representation of this protein with one of the surface depicted (right panel).
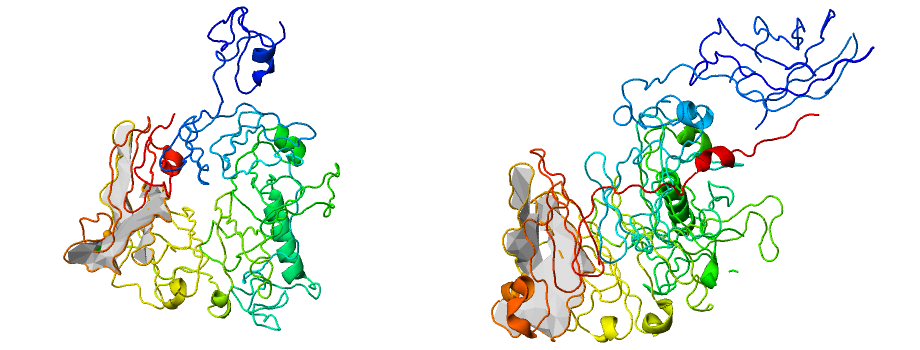
Fig. 9 Example of proteins with wrong lasso topology (L3,1 and SL4) but high GDT score (GDT >88) predicted during CASP competition.
Intelligent design of engineered disulphide bridges in proteins and other biopolymers, medicine
Yet another application is to design proteins with a new topology, e.g. via disulphide bridges, which are commonly used in protein engineering [8].
- It is well known that engineering cysteine bonds “is considered to be a straightforward approach to 'trap' a protein in a particular conformation or increase proteins stability” [7,9]. Additionally the web server 'Disulfide by Design 2' is a helpful tool to predict the placement of new cysteines to increase the stability of protein structures [10]. Connection of this server with LassoProt server can be used to identify a correlation between topology and stability of proteins. Such an approach was used already in [8] for a few proteins with the alpha helical bundle fold.
- The server can be used to augment known structures. E.g. cysteine knot peptides were used by the Cochran group to engineer a new class of high affinity molecular imaging agents [11], based on the konttins proteins (characterized by a cystine knot introduced into a small peptide). Similar approach could be used based on the complex lasso motif. Moreover, the server can be used to design proteins with desired topological motif, which can be used as scaffolds in medicine, similarly as it was performed in the case of the cyclotides [12].
- The LassoProt server can be used to identify introduced “unwanted” topology [7]. E.g. “artificial” lasso L1,1 was found in the crystal structure (pdb code 2YHG) of the recently de novo designed nucleoside hydrolase and it is shown in the Fig. 10.
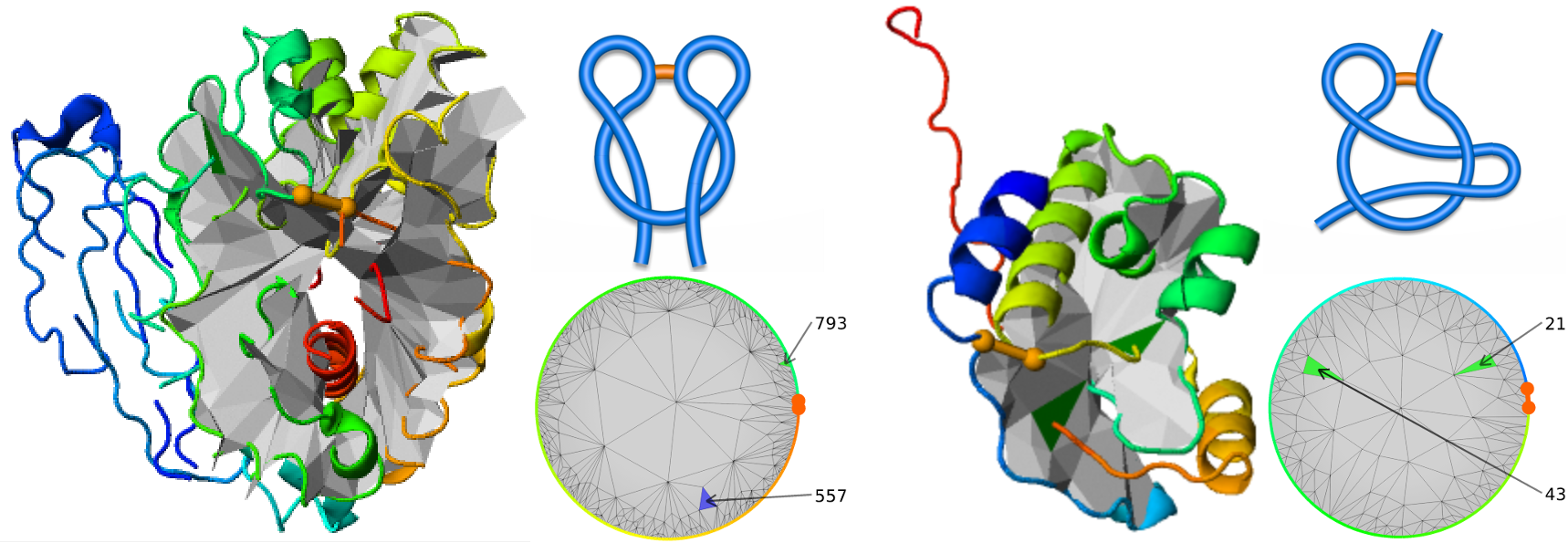
Fig. 10 Two proteins deposited in the pdb with “artificial” lasso. Left, the protein (pdb code 2YHG) with “artificial” double lasso L1,1. Right, conserved hypothetical protein (pdb code 1ZD0) with “artificial” supercoiling topology LS1.
Mathematics, detection of topology
Yet another application is provided via fast and flexible way to detect other complex topologies, where a minimal surface method could be applied. E.g. our server can be used to analyze so called tadpoles [13] or complex topology designed chemically, such as shown in the Fig. 11 – based on [14].
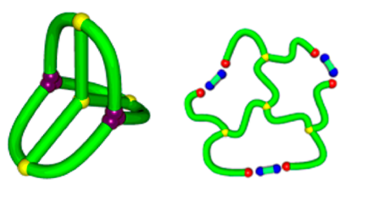
Fig. 11 Example of chemically constructed tetracyclic polymer topology in the group of Tezuka [14].











