A server and database of proteins with lassos
LassoProt collects information about proteins and other (bio)polymers with lassos [1]. LassoProt detects covalent loops (closed by cysteine, as well as amide, ester, thioester and other bridges) in a given (bio)polymer, determines surfaces of minimal area spanned on such loops, and analyzes if and how termini of the polymer pierce these minimal surfaces (introduced in [2]). Each lasso motif, can be visualized directly on an interactive protein scheme or its simplified version (JSmol), as well as on a barycentric representation of the minimal surface. This permits users to determine e.g. piercing residues, depth of a lasso (length of lasso tails), or the area of lasso loop. In addition, the database presents comprehensive information about the biological function of proteins with lasso motifs, their families, the organisms and fold types of these proteins. The database is updated each week (based on pdb automatic scan), and currently deposited data is summarized in database statistics.
LassoProt server allows users to submit proteins, polymer or polygon structures, or a set of chain configurations (e.g. data from molecular dynamics and Monte Carlo simulations, uploaded as entire trajectories), and generate their lasso fingerprints. Moreover, lasso-type entanglement can be automatically detected based on cysteine, amide, ester or thioester bridges, or on a closed loop designed by the user. Each determined lasso is supported with a set of data files enabling users their own analysis and visualizing results in other programs, such as Mathematica or vmd. The simplified chain with minimal surface is also provided.
Lasso fingerprints determined by our server have broad applications [3,4,5,6], ranging from applied mathematics, through (bio)physics, (bio)chemistry, and protein structure prediction, up to analysis of biological functions of proteins and medicine. Examples of applications of the server are provided in Apply results of lasso topology analysis.
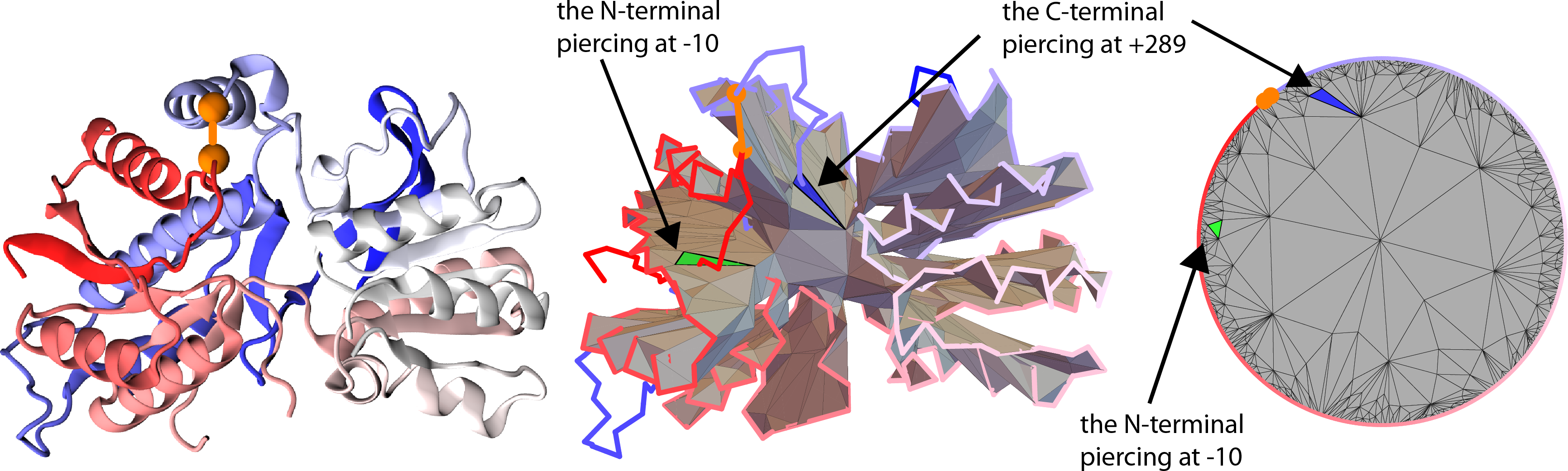
Fig. 1 Glutamate receptor with pdb code 3OM0 in its cartoon representation, along with the minimal surface prescribed for it and barycentric graph (based on [2]).
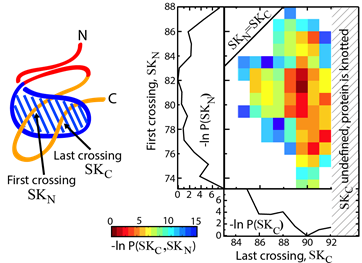
Fig. 2 Estimated effective potential (PMF) along the loop crossing coordinate for the smallest knotted protein (pdb code 2EFV). The left panel shows a schematic representation of a surface spanned on a twisted loop of the knotted protein, which is pierced via the C-terminus twice (double lasso - L2). The right panel shows tracked behavior of residues during crossing the plane of the loop based on the explicit-solvent simulation [6]. The crossing data are used to construct an effective potential to describe knotting mechanism.
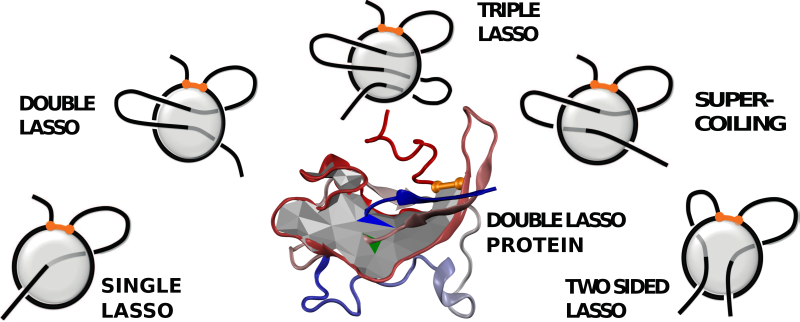
Fig. 3 Different classes of lasso-type entanglement and the example of complex lasso structure (in the center).
[1] Dabrowski-Tumanski P, Sulkowska JI (2016) Unique properties of lasso proteins. - under review
[2] Niemyska W, Dabrowski-Tumanski P, Kadlof M, Haglund E, Sułkowski P, Sulkowska JI (2016) Complex lasso: new entangled motifs in proteins.
[3] Haglund E, Sulkowska JI, He Z, Feng GS, Jennings PA, Onuchic JN (2012) The unique cysteine knot regulates the pleotropic hormone leptin. PLoS One.7(9):e45654
[4] Haglund E, JI Sulkowska, JK Noel, H Lammert, JN Onuchic, PA Jennings (2014) Pierced Lasso Bundles are a new class of knot-like motifs. PLoS Comput Biol. Jun 19;10(6):e1003613
[5] Haglund E (2015) Engineering covalent loops in proteins can serve as an on/off switch to regulate threaded topologies. J. Phys. Cond. Matter 27, 35
[6] Noel JK, Onuchic JN, Sulkowska JI (2013) Knotting a Protein in Explicit Solvent. J. Phys. Chem. Lett. 4 (21), pp 3570–3573
Authors
This server and database has been created in a joint collaboration between: Paweł Dąbrowski-Tumański (University of Warsaw, Faculty of Chemistry and Centre of New Technologies), Wanda Niemyska (University of Warsaw, Centre of New Technologies and University of Silesia, Institute of Mathematics), Paweł Pasznik (University of Warsaw, Centre of New Technologies) and Joanna I. Sulkowska (University of Warsaw, Faculty of Chemistry and Centre of New Technologies).
Funding
The research leading to creation of this database has been supported by: National Science Center [grant agreement Sonata BIS 2012/07/E/NZ1/01900 to J.S.]; Foundation for Polish Science [SKILLS/Inter grant agreement 130/UD/SKILLS/2015 co-financed by European Social Fund to W.N.].



