Introduction
The LassoProt is a server detecting any type of lasso entanglement (in a chain with at least one free terminus) both in single frames and whole trajectories and a database collecting information about complex lasso proteins [1]. The lasso entanglement arises, when one terminus of a protein backbone pierces through an auxiliary surface of minimal area, spanned on a covalent loop (see Fig. 1).
The LassoProt server is designed to study protein chains, however one can upload any kind of (bio)polymer. It is equipped with various tools facilitating perceiving the lasso entanglement. In particular, the user can visualize the protein with the minimal surface spanned on a covalent loop and in trajectory analysis LassoProt offers two dynamical charts describing the change in topology during the trajectory
The database classifies protein chains first according to the type of chemical interaction [2], which form a covalent loop, second to its entanglement type i.e. number and the direction of threading. Based on these results, the lasso type is prescribed to every closed loop and the overal topology (lasso fingerprint) to the protein chain. For further analysis, LassoProt also presents many biological and geometrical statistics. The database contains all the protein structures deposited in Protein Data Bank and is automatically updated every Wednesday.
The introduction section contains two parts:

Fig. 1 Schematic representation of a complex lasso protein.
Server overview
The first example of complex lasso protein was leptin, in which the loop is closed via cysteine bridge [3-5]. This premier discovery was followed by the thorough study of the non-redundant set of protein chains containing the cysteine-bridge-based loop [1]. This analysis revealed the existence of five distinct major classes, however other classes are possible especially for chains with large gap (treated by us as Artifacts), or by deposition of newly cristallized chains. The most popular class are the single lasso proteins (L1), in which a covalent loop is pierced by a tail only once. The next are double (L2) and triple (L3) lassos, in which the covalent loop is pierced twice or three times respectively, while after piercing the tail winds back and performes next piercing from the opposite direction. The two remaining classes are two sided-lassos (LLi,j) in which both tails cross the surface i and j times respectively and supercoiling lassos (LS) in which one tail winds around the covalent loop after crossing and then performes piercing from the same direction. These major classes are schematically depicted in Fig. 2. We stress that complex lasso proteins with cysteine loops should not be confused with well known cysteine knot proteins and knottins [6] nor knotted proteins [7]. The complex lasso motif is more general notion than cysteine knots, as cysteine knots require the existence of at least three cysteine loops.
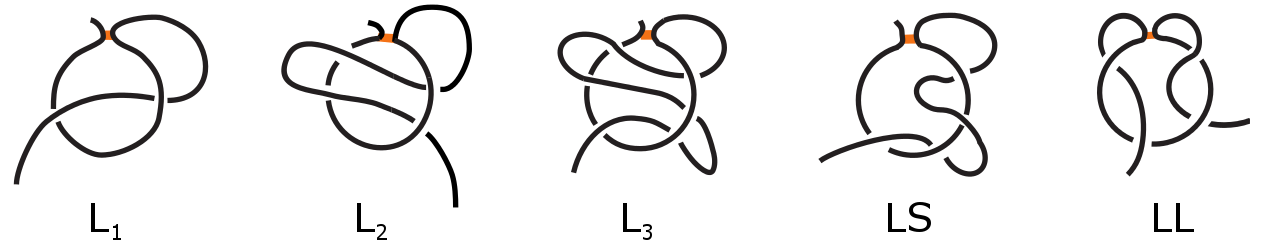
Fig. 2 Major classes of complex lasso proteins: single lasso L1, double lasso L2, triple lasso L3, supercoiling LS2 and two-sided lasso LL1,1.
Although the cysteine-bridge-based complex lasso proteins are the most abundant ones, LassoProt detects and classifies also the proteins with stable chemical bridge based on C-N, C-O, C-S (e.g. amide, ester or thioester) and other bonds (see Type of lasso section). Thus LassoProt first identifies the chemical nature of the a closed loop, then assigns a lasso type based on the number and direction of piercings. We would like to stress, that the same analysis revealing the entanglement motif with respect to closed loops can be applied to artificially designed proteins or other (bio)polymers, e.g. DNA or RNA via the LassoProt server. Moreover, understanding of entangled protein properties requires analysis of its dynamical behaviour. Therefore, the LassoProt server is equiped with the ability to study whole trajectories.
Usually, determination of the threading is impossible by a naked eye. Therefore, we developed [1] the mathematically well defined method which specifies the piercings with its direction unequivocally (see more in the Lasso detection section). This on the other hand enables us to split every major class into smaller subclasses, according to the piercing tail (N- or C-terminal) and the direction of piercing. Then, the proteins can be classified according to the class of its covalent loops (see Lasso type classification section).
There are three main options a user can choose from to view or analyze data, which are visible in the main page, or in the top of the site (Fig. 3):
- browse database, enabling a user to browse all structures currently deposited in the database;
- search database, enabling a user to search and classify proteins based on topology, geometry, or different biological and sequential properties;
- process my structure, allowing the upload of new proteins, polymer-like structures or simulation trajectories and analyze their lasso type (and its time evolution in case of trajectory).
Fig. 3 Main page of LassProt Database with the main options in the top.
After choosing or uploading some particular protein (or any other polymer chain), its lasso entanglement motif for closed loop (via S-S, C-N, C-O, C-S interaction or any other closed loop chosen by the user) is presented. Each protein chain existing in database is supplied also with the biological information comprising data from other databases (pubmed, rcsb, pfam and doi for article of reference, if exists). To fully understand the topological features of the protein, we encourage users to utilize also the KnotProt server in which the knotted topology of a polymer can be checked.
The posttranslational modification joining the side groups is known to introduce greater stability to the protein structure. Moreover, it is well known that cysteine bridges are important for biological function of proteins. More information about usability of the data gathered in the database or obtainable via the server are contained in the "Apply results" section. E. g. data included in the LassoProt show that a local constraint (not only imposed by cysteine bridges) has a consequence for the global structure of proteins. Therefore, the complex lasso entanglement is expected to play an important role for proteins. However, the function, and the properties can depend on the lasso type, thus a detailed representation of protein geometry is crucial for understanding their role (see Interpreting lasso data). The LassoProt is designed to facilitate searching the possible correlations between topology/geometry and biological properties. We believe, that the server will help researchers to identify and understand the influence of geometrical constraints on biological function, stability and structures of biolymers.
To provide users a broad spectrum of ways the LassoProt can be used, it contains detailed information about the geometry of every closed loop in the protein chain. Moreover, it provides extensive statistics about complex lasso proteins based on their biological function, molecular tags, family association, type of fold, as well as geometric data: tails and closed loops lengths, piercings positions etc. The ability to upload own structures and trajectories allow to study e.g. folding pathway in proteins, where the lasso type can serve as a new reaction coordinate. The other applications are presented in "Apply results" section. However, because the user can design the chain or its topological constraints himself, the LassoProt server is fully adjustable providing unique tools to go beyond the results and applications presented here.
The LassoProt database/server is a useful and easy to use tool designed to analyze the new entangled motif - lasso. However, we are aware that we could not foresee every possible need of potential user. Therefore any remarks concerning the database as well as ideas of introducing new utilities are most welcome.
[1]
Niemyska W, Dabrowski-Tumanski P, Kadlof M, Haglund E, Sułkowski P, Sulkowska JI (2016) Complex lasso: new entangled motifs in proteins.
[2] Dabrowski-Tumanski P, Sulkowska JI
Unique properties of lasso proteins. - under review
[3] Haglund E, Sułkowska JI, He Z, Feng GS, Jennings PA, Onuchic JN (2012)
The unique cysteine knot regulates the pleotropic hormone leptin. PLoS ONE 7:e45654.
[4] Haglund E, Sulkowska JI, Noel JK, Lammert H, Onuchic JN, Jennings PA (2014)
Pierced Lasso Bundles Are a New Class of Knot-like Motifs. PLoS Comput Biol 10:e1003613
[5] Haglund E. (2015).
Engineering covalent loops in proteins can serve as an on/off switch to regulate threaded topologies. J. Phys. Condens. Matter, 27:354107.
[6]
The Knottin Database
[7] Jamroz M, Niemyska W, Rawdon EJ, Stasiak A, Millett KC, Sułkowski P, Sulkowska JI (2014)
KnotProt: a database of proteins with knots and slipknots. Nucleic Acids Research
Protein chains in the database
The database contains almost every protein chain deposited in the Protein Data Bank - redundant chains within particular pdb entry of homomultimeric complex and highly homological sequences are represented by one chain only. However, because the lasso type may vary upon change of a single, crucial, bridge-forming amino acid, or oxidation potential (in case of S-S bond) we treat the chains as identical, if they are sequentially identical and they represent the same lasso motif (more about defining lasso type is presented in Determination of lasso type section). The database is self-updating based on new pdb entries each week.
Examples:
- Change of lasso type based on chemical condition
One of the examples can be the Glutamate receptor 2. The structure crystalized in oxidizing condition (with
pdb code
3T9U) has the overal lasso pattern (fingerprint) L
2L
1, while its reduced form (with
pdb code 3T9V) has no cysteine bridges and therefore has a trivial topology.
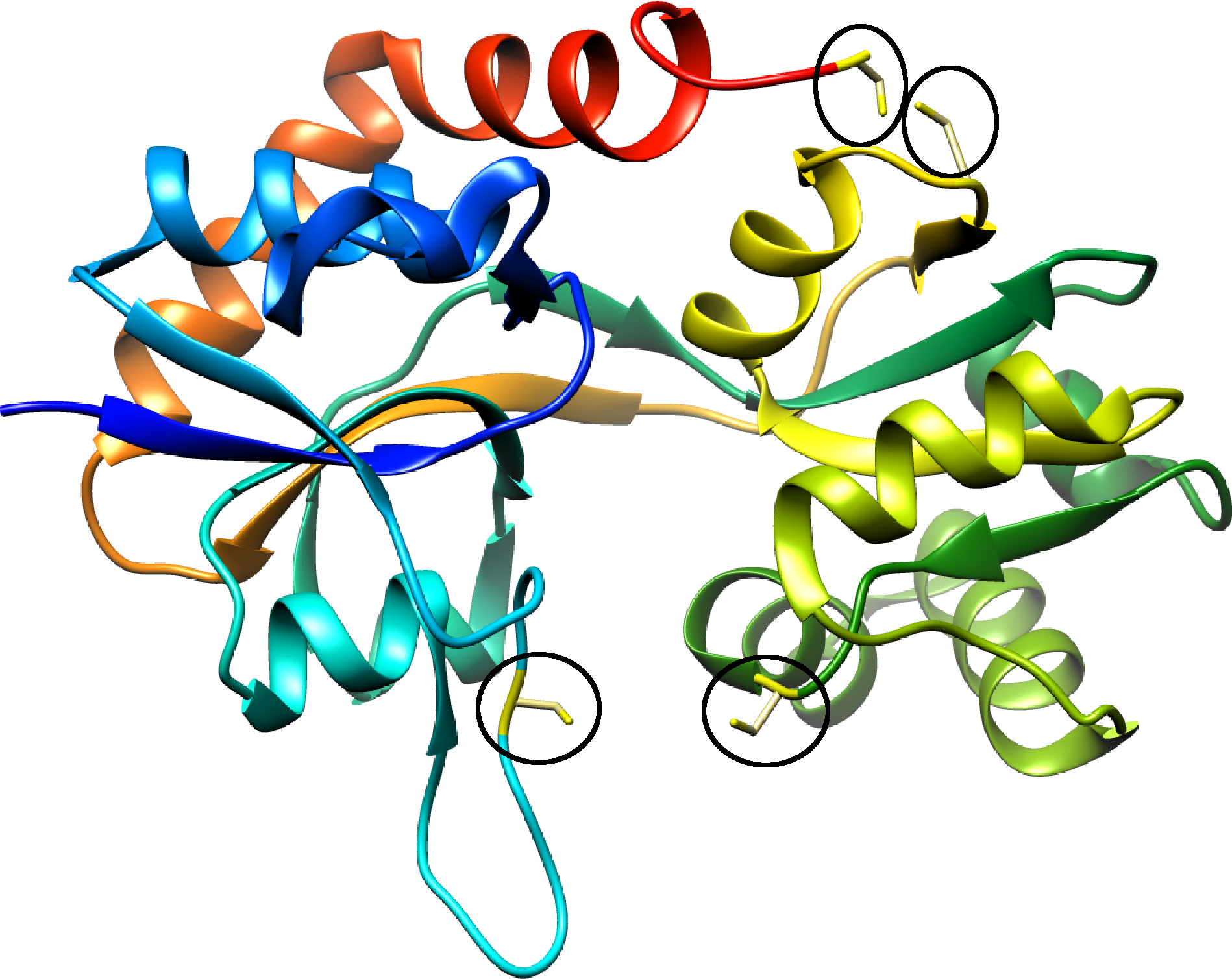
Fig. 4 The crystal structure of Glutamate receptor 2 with pdb code 3T9V. The black ovals highlight the reduced cysteines, which should form bridge in order to introduce non-trivial entanglement.
- Change of lasso type based on single amino acids substitution
Glutamate receptor is a good example of the influence of amino acids substitution on the lasso type of the chain. Its native structure with
pdb code
4FAT has the overal lasso pattern (fingerprint) L
1. However, there exists a Glutamate receptor mutant, which has two amino acids Ala63 and Ser140 changed to bridge-forming cysteins. The bridge introduced changes the lasso type of the protein to L
2L
1 in Glutamate receptor with
pdb id
3T9V.
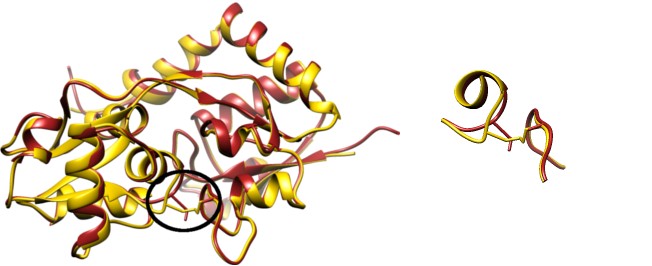
Fig. 5 The crystal structure of Glutamate receptor 2 - left, overlayed structures with pdb code 4FAT (dark red) and 3T9V (yellow). The black oval shows the region with introduced cysteine bridge. The same region is enlarged in the right panel.
PDB files
Apart from standard X-ray structures, we include non-X-ray entries and entries with Cα atoms only. Chains are subsequently evaluated to take into account existence of non-typical aminoacids: CIT, HEY, HYP, ORN, SEC, PYL, ASX, GLX, XLE, XAA, MSE, FGL, LLP, SAC, PCA, MEN, CSB, HTR, PTR, SCE, M3L, OCS, KCX, SEB, MLY, CSW, TPO, SEP, AYA, TRN, and D-amino acids: DAL, DAR, DSG, DAS, DCY, DGN, DGL, DHI, DIL, DLE, DLY, MED, DPN, DPR, DSN, DTH, DTR, DTY, DVA. This analysis is performed so as not to introduce additional breaks along protein chain. In case of NMR structures, we take the first model with a given chain name.
Gaps
The gaps in the structure are “modeled” as a straight segment. This can however change the lasso type of the protein. Therefore, if the gap is larger than 6 residues and our analysis results in non-trivial topology of the closed loop, the chain is classified as an artifact, so one should be careful in interpreting results in such cases. In order to provide users with the most accurate data, we plan to model the missing parts of the proteins in the future.
Modelling the gap of a length less than or equal to 6 residues as a straight line should not change the overall topology of the protein chain. However, in each case the user is warned about breaking of the chain. Missing atoms in the chain are denoted in sequence representation as "-" and pdb code is denoted by the sign .





